Background
Centralized support for genomic methods and bioinformatic data analysis is a vital component of our collaborative research consortium. To ensure success, this support must be paired with continuous development of state-of-the-art methods that keep pace with the evolving needs of the research.
Strategy
We provide essential expertise and have established advanced methods in genomics and bioinformatics. These will be specifically tailored to support CRC research projects, enabling crucial insights into CNS recovery checkpoints. In this project, we are offering central support, creating a robust training infrastructure, and conducting original research focused on method development and integrative data analysis.
Z02 Genomics and Bioinformatics Platform
Since the first funding period, Z02 has provided comprehensive support for all our research projects, from experimental design to bioinformatics data analysis. We specialize in bulk and single-cell genomic methods, including technologies like 10x Genomics and flow-cytometry based isolation methods. Additionally, we offer training on tissue preparation, single-cell sorting, and the use of 10x Genomics equipment to ensure high-quality data.
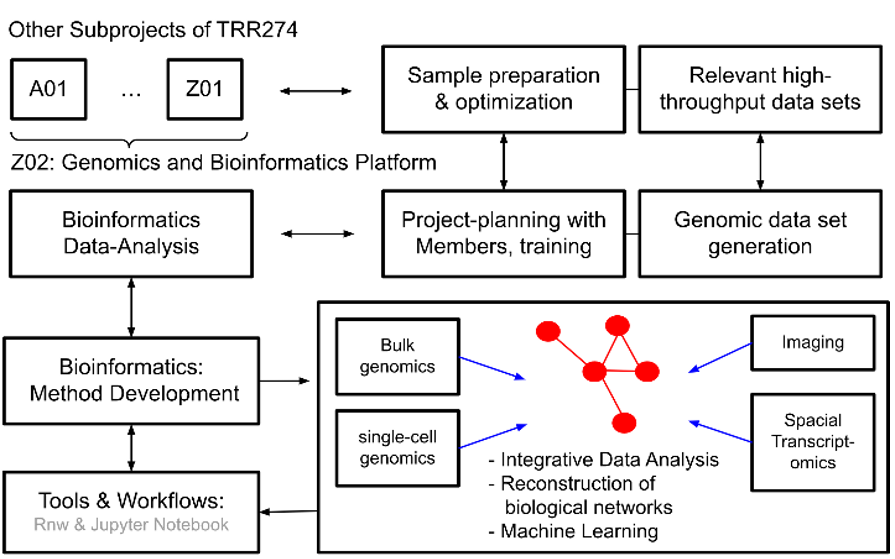
By centralizing sample preparation, sequencing, and analysis, our platform ensures quick turnaround times and reliable data. Our work has led to important discoveries, such as identifying reactive glial states that drive pathological processes in injury conditions.
In the next funding period, we aim to expand our methods and tools, particularly focusing on integrating data across different models and species. This will involve combining various types of data, including genomic, epigenomic, transcriptomic, and imaging data, to create standardized analysis pipelines.
Our Key Goals:
Want to collaborate?
Find detailed and relevant information on our password-protected internal page.





